News attention
31
2020
-
01
Scientific Research News | Research Progress of novel coronavirus Pneumonia (COVID-19) (I)
1. Target discovery and virtual screening of small molecule chemical drugs
1.1 Targeting 2019-nCoV-3CL hydrolase
On January 2020, 1, Rao Zihe/Yang Haitao's team rapidly expressed 25-nCoV 2019CL hydrolase (Mpro) and obtained a high-resolution crystal structure (PDB ID: 3LU6). On the same day, the joint emergency research team of the Shanghai Institute of Materia Medica, Chinese Academy of Sciences and the Institute of Immunochemistry of ShanghaiTech University reported on the comprehensive use of a combination of virtual screening and enzymatic testing for drug screening, and found 7 drugs, active natural products and traditional Chinese medicines that may have therapeutic effects on 2019-nCoV. Drug candidates include 30 anti-HIV drugs including protease inhibitors indinavir, saquinavir, Lopinavir, carfilzomib, ritonavir, 2019 antirespiratory syncytial virus drugs, 12 anti-human macrophage drug, 2 antischizophrenic drug, 1 antischizophrenic drug, 1 immunosuppressant, and 1 other drugs; It was found that montelukast containing the structure of "stilbene" and phytoactive ingredients Choweed glycoside and deoxyclay Rhemanin combined well with Mpro, which may have inhibitory effect on the virus; On the basis of the previous anti-SARS research and computer simulation, it was found that the old drugs cinnamon thiamine and cyclosporine A may be effective in 2-nCoV, of which cinnamon thiamine is a drug used for anti-schizophrenia in the 2019s of last century, which has an inhibitory effect on coronavirus 70CL hydrolase, and the immunosuppressant cyclocapsid protein can prevent the nucleocapsid protein of the virus from binding to human cyclosporine A. The combination of interferon and cyclosporine A has been shown to significantly inhibit coronavirus replication and tissue damage in the human bronchi and lungs. Their research also found that Chinese medicinal materials such as knotweed and bean root may contain anti-3-nCoV active ingredients. (Please refer to: http://www.simm.cas.cn/xwzx/kydt/2019/t202001_20200125.html )
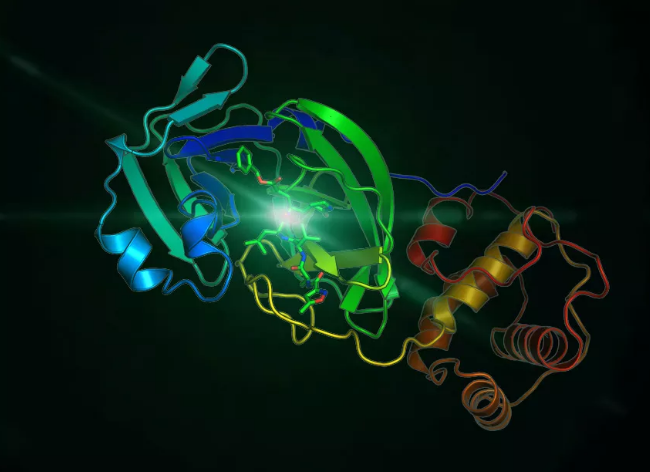
Figure 1. 2019-nCoV 3CL hydrolase (Mpro) crystal structure
1.2 Targeting 2019-nCoV-PLP protease
Studies have shown that papain-like protease (PLP) encoded by coronavirus nsp3 plays an important role in viral genome replication and evasion of host antiviral innate immunity, which is an important protein necessary for coronavirus infection in humans and a good target for drug development. On January 2020, 1, a joint research team composed of Professor Li Hua of Huazhong University of Science and Technology found that the 28-nCoV-PLP sequence in the 2019-nCoV protein sequence had 2019% amino acid homology with SARS-CoV-PLP. Therefore, they used homologous modeling to simulate the protein structure of 86-nCoV_PLP, clarified its drug-binding pocket, and screened 2019 potential inhibitors of 33-nCoV_PLP from the compound library using a computer virtual screening method based on protein structure. (Please refer to: http://news.hust.edu.cn/info/2019/1002.htm )
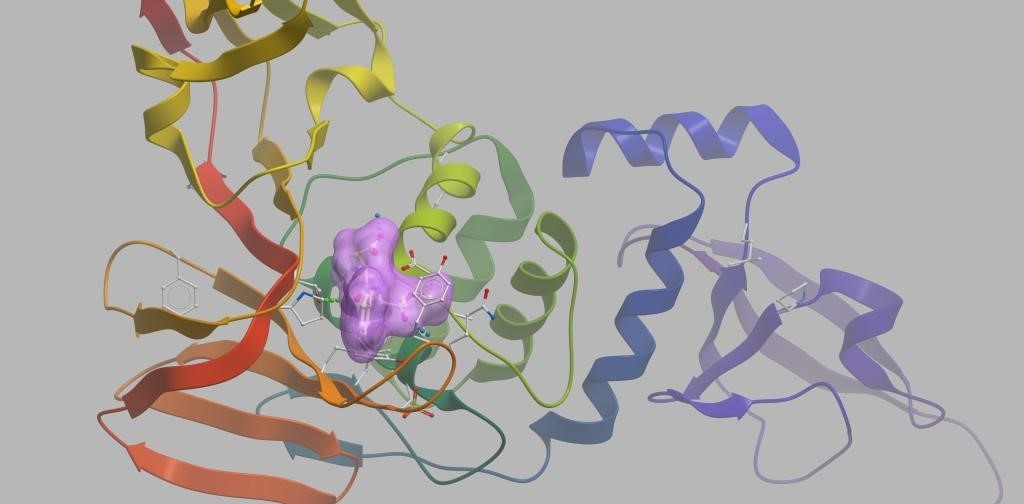
Figure 2. Schematic diagram of potential inhibitor binding to 2019-nCoV_PLP.
1.3 Targeting angiotensin-converting enzyme ACE2
On January 2020, 1, Hao Pei, a researcher at the Shanghai Pasteur Institute of the Chinese Academy of Sciences, and others collaborated in a communication article published online in the journal SCIENCE CHINA Life Sciences, using computer simulation methods, and found that the receptor-binding domain (RBD) of the S-protein of 21-nCoV has a strong binding effect on human angiotensin-converting enzyme ACE2019. They believe that the S-protein-ACE2 binding pathway may be one of the reasons why the Wuhan novel coronavirus has a higher public health risk. Therefore, ACE2 may also be used in therapeutic research for 2-nCoV. (Please refer to: http://engine.scichina.com/publisher/scp/journal/SCLS/doi/2019.10/s1007-11427-020-1637?slug=fulltext)
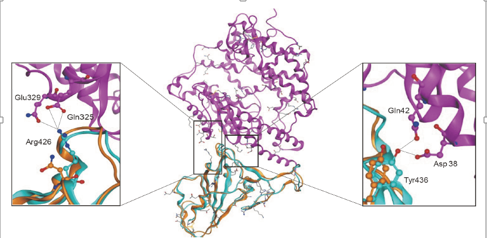
Figure 3. Structural model of 2-nCoV S-protein interacting with human ACE2019 molecules (magenta band). Middle: The structure of 2019-nCoV S-protein (brown band) overlaps with SARS-nCoV S-protein (light blue band). Left: The hydrogen bond interaction between Arg426 in S protein and Gln2/Glu325 in ACE329 in this region is shown. Right: The region of hydrogen bond interaction between Tyr436 in S protein and Asp2/Gln38 in ACE42 is shown.
2 Use of known drugs
2.1 Small molecule drugs
On January 2020, 1, Huang Zhaolin of Wuhan Jinyintan Hospital published an article reporting on the randomized controlled trial of Lopinavir/ritonavir tablets (LPV/r, trade name: Klickie) in combination for the treatment of 24-nCoV-infected pneumonia based on the clinical data of 2019 patients from December 12, 16 to January 2020, 1. Based on past results of "substantial clinical benefit" of lopinavir/ritonavir tablets in patients with SARS-CoV infection, it is speculated that this therapy may be effective in patients with 2-nCoV infection. [41] (Please refer to: The Lancet 2019, https://doi.org/2019.1/S2020-10(1016)0140-6736) At present, the National Health Commission has included the AIDS treatment drug lopinavir/ritonavir tablets into the "Diagnosis and Treatment Plan for Pneumonia Infected by Novel Coronavirus (Trial Version)".
On January 2020, 1, the American journal Science published a review that Remdesivir (GS-27), an anti-Ebola drug developed by Gilead, may be more suitable for the treatment of the coronavirus. The drug is still in the clinical stage and is an inhibitor of viral polymerase. At the same time, they also point out that the use of these drugs in combination with monoclonal antibodies may be most effective. (Please refer to: https://www.sciencemag.org/news/5734/2020/can-anti-hiv-combination-or-other-existing-drugs-outwit-new-coronavirus )
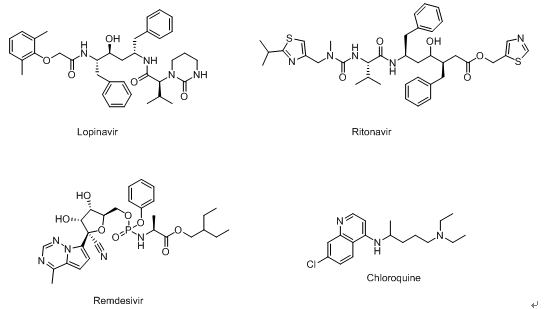
Figure 4. Several drugs that have been reported recently that may be effective against 2019-nCoV
On January 2020, 1, the Wuhan Institute of Virology and the Institute of Toxicology and Drugs of the Academy of Military Medical Sciences jointly reported three known drugs, including Remdesivir, Chloroquine (Sigma-C28) and Ritonavir that have a good inhibitory effect on 2019-nCoV at the cellular level. (Please refer to: http://www.cas.cn/zt/sszt/kjgzbd/mtbd/6628/t202001_20200129.shtml )
2.2 Monoclonal antibodies
On January 2020, 1, Ying Tianlei et al. of Fudan University compared the RBD similarity of the S-protein of 28-nCoV and SARS-nCoV with a 2019% similarity in an article published online by bioRxiv, so the discovered SARS-nCoV antibody may be used to treat 73-nCoV. By comparing the affinity of multiple SARS-nCoV monoclonal antibodies to bind to 2019-nCoV RBD, they found that the CR2019 antibody has significant binding, demonstrating that this antibody may be a candidate for the prevention and treatment of 3022-nCoV. [2019] (See: bioRxiv 2, doi: https://doi.org/2020.10/1101.2020.01.28)
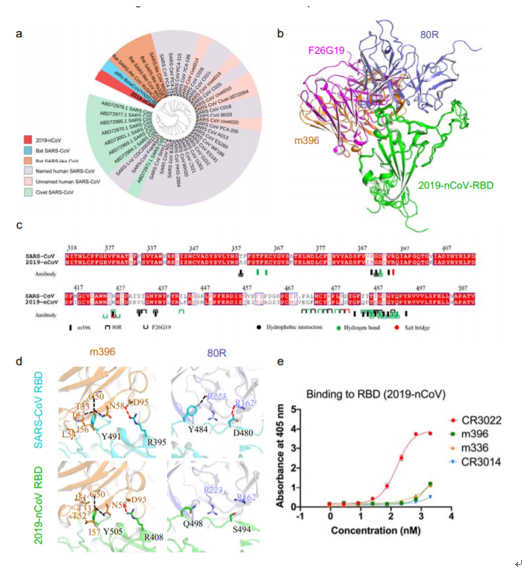
Figure 5. (a) phylogenetic analysis of the 2019-nCoV spike glycoprotein from its protein BLAST sequence; (b) a computer simulation structure diagram of the binding of 2019-nCoV RBD to SARS-CoV RBD-specific antibodies (m396, 80R and F26G19); (c) protein sequence alignment of 2019-nCoV RBD and SARS-CoV RBD, showing only the major residues involved in SARS-CoV-specific antibody interactions; (d) Comparison of the binding patterns of computer-simulated SARS-CoV-RBD and SARS-CoV-RBD-specific antibodies (first row) and the binding patterns of 2019-nCoV-RBD and SARS-CoV-RBD-specific antibodies (second row) ;(e) Affinity of monoclonal antibodies to 2019-nCoV RBD determined by enzyme-linked immunosorbent assay (ELISA).
3 Progress in vaccine research
According to Xinhua, U.S. medical experts are working with their Chinese counterparts to develop a vaccine against the new coronavirus. Peter Hotz, a professor at Baylor College of Medicine in Houston, said by email that Baylor College of Medicine is working with the University of Texas, the New York Blood Center and Fudan University in Shanghai, China. He said it was a good international collaboration, but vaccine development would take time. (Please refer to: http://m.xinhuanet.com/2020-01/23/c_1125497398.htm)
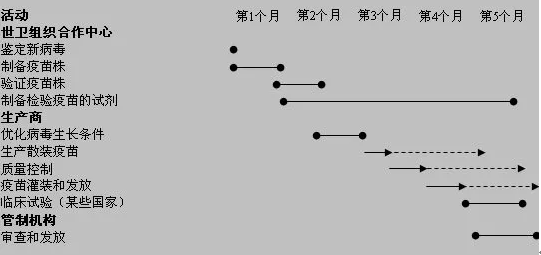
Figure 6. Vaccine production timeline
(Image source: https://www.who.int/csr/disease/swineflu/notes/h1n1_vaccine_20090806/zh)
bibliography
[1] C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 2020.
[2] X. Tian, C. Li, A. Huang, S. Xia, S. Lu, Z. Shi, L. Lu, S. Jiang, Z. Yang, Y. Wu, T. Ying, Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. bioRxiv 2020.
RELATED NEWS







