News attention
24
2020
-
04
Scientific Research News | Research Progress of novel coronavirus Pneumonia (COVID-19) (49)
■ On April 4, Carly G. K. Ziegler of Harvard Medical School et al. published an online report titled "SARS-CoV-22 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across." tissues", which utilizes human, non-human primate, and mouse single-cell RNA sequencing (scRNA-seq) datasets across health and disease datasets to discover putative targets for SARS-CoV-2 in tissue-resident cell subsets.
This study identified ACE2 and TMPRSS2 co-expression cells in type II lung cells, ileal resorptive intestinal epithelial cells, and nasal goblet secretory cells. Surprisingly, the study found that ACE2 is an interferon-stimulated gene (ISG) in airway epithelial cells. The study data suggest that SARS-CoV-2 can enhance infection using species-specific interferon-driven ACE2 upregulation.
There are three different types of interferon: type I interferon, type II interferon, and type III interferon. Each appears to focus on almost indistinguishable reactions mediated by the binding of STAT1 homodimers or STAT1/2 heterodimers to ISG. However, there is growing evidence that each type of IFN may have a non-redundant role in host defense or immunopathology, particularly on the epithelial barrier.
Studies have shown that the timing of the type I IFN reaction is a key component of the reaction in vivo. In the absence of controlled trials targeting mixed-effects, clinical treatment of SARS-CoV, MERS-CoV, and SARS-CoV-2 using approved IFNs has been attempted, and symptoms have been found to improve or worsen rapidly, elucidating specific ISGs of tissue and cell types and their activity is critical to understanding IFN's role in host defense during SARS-CoV-2 infection in humans[1].
■ On April 4, Liang Tingbo's team from Zhejiang University published a report entitled "Viral load dynamics and disease severity in patients infected with SARS-CoV-21 in Zhejiang province, China, January-March 2: retrospective cohort" online in BMJ study", which systematically evaluated 2020 patients (2 mildly ill and 96 severe) infected with SARS-CoV-22 after admission to hospital; from 74 January 2020 to 1 March 19) and analyzed temporal variations in viral load and correlations between viral load across different sample types and disease severity. The study found that RNA was detected in the stool of 2020 patients (3%) and in the serum of 20 patients (3000%). One patient tested positive for SARS-CoV-55 in a urine sample. The duration of virus in stool (59 days, interquartile range 39-41 days) was significantly longer than in respiratory (2 days, 22-17 days) and serum samples (31 days, 18-13 days). The duration of the virus in respiratory samples of patients with severe disease (29 days, 16-11 days) was significantly longer than in patients with mild disease (21 days, 21-14 days). In the mild group, viral load in respiratory samples peaked from the second week of disease onset, while in the severe group, viral load continued to rise in the third week. The virus lasts longer in patients over 30 years of age and in male patients.
Overall, the duration of SARS-CoV-2 in stool samples was significantly longer than in respiratory and serum samples, highlighting the need for enhanced management of stool samples in the prevention and control of the epidemic, and the persistence of the virus in respiratory tissues for longer periods of time and greater loading, with a later peak in critically ill patients [2].
■ On April 4, the team of Li Lanjuan, academician of the Chinese Academy of Engineering, published a research paper "Patient-derived mutations impact pathogenicity of SARS-CoV-19" online on the medical preprint platform medRxiv. The paper pointed out that the new coronavirus has appeared mutations that can actually affect pathogenicity, and its mutation and diversity may be greatly underestimated; Different variants can vary up to 2-fold in cytopathic effects and viral load.
The research team isolated 11 strains of the new coronavirus from patients admitted to the First Affiliated Hospital of Zhejiang University. The patients ranged in age from 4 months to 71 years, and 10 of them had a history of exposure to Wuhan. After ultra-deep sequencing of 11 isolated strains on the Novaseq 6000 platform and comparison with 1111 genome sequences from the GISAID database, it was found that the 11 patients isolated a total of 33 mutations in the virus, 19 of which were new mutations identified for the first time.
The researchers also infected Vero-E6 cells with the isolated strains, quantitatively assessing their viral load at 1, 2, 4, 8, 24 and 48 hours post-infection, as well as viral cytopathic effects at 48 and 72 hours post-infection. Among them, Vero-E6 cells (African green monkey kidney cells) are a kind of alloploidy cells, and the famous Hela cell line and canine kidney cells (MDCK cells) are commonly used cell lines. This cell line has an ACE2 receptor that is very similar to human cells.
It was found that when infecting Vero-E6 cells, the observed mutations had a direct impact on viral load and cytopathological effects, with up to 270-fold differences. The higher the viral load, the higher the lesion effect and mortality of cells. This finding suggests that the viral mutations observed in the study can significantly affect the pathogenicity of the new coronavirus, and such mutations may be present in virus strains collected around the world [3].
■ On April 4, the international academic journal Science Advances published the latest results of the Liu Shi research group of the State Key Laboratory of Virology in the form of a cover paper, entitled "O-GlcNAc Transferase Promotes Influenza A Virus-Induced Cytokine Storm by Targeting Interferon Regulatory." Factor-17”。 This work identifies a pair of molecular targets that interact with inflammation and glucose metabolism during influenza virus infection and analyzes their regulatory mechanisms.
Glucose metabolism is the most important way of intracellular energy metabolism, and viral infection induces inflammation response must require energy supply. Does the glucose metabolism signaling network interact with the inflammatory signaling network? What is the mechanism? These questions become an important scientific question in the field of virology that has not yet been elucidated. O-GlcNAc transferase (OGT) is a mammalian essential enzyme that attaches N-acetylglucosamine (GlcNAc) to the serine and threonine hydroxyl groups of proteins in β-configuration O-glycosidic bonds. The post-translational modification of this protein has a regulatory effect on many signaling pathways within cells and is closely related to the occurrence and progression of a variety of major diseases. In this study, OGT was found to play an important role in influenza virus-induced cytokine storms through animal and cellular models. Subsequent mechanistic studies have shown that during influenza virus infection, OGT attaches GlcNAc to IRF5 by interacting with IRF5. Glycosylated IRF5 subsequently recruits TRFA6 and induces IRF5 ubiquitination. The study revealed that reprogramming cellular metabolic activity plays a key role in immune system activation and excessive inflammation, and also provides new clues for screening antiviral drugs [4].
■ After two months of efforts, the research team of the establishment of a new coronavirus (SARS-CoV-2) animal model led by the Institute of Medical Biology of the Chinese Academy of Medical Sciences and the National Kunming High-level Biosafety Primate Experimental Center Peng Xiaozhong, Liu Hongqi and Lu Shuaiyao and the team of Ke Changwen of the Guangdong Provincial Center for Disease Control and Prevention successfully established a non-human primate infection model of SARS-CoV-2. On April 2, the research results were published in the bioRxiv preprint under the title "Comparison of SARS-CoV-4 infections among 17 species of non-human primate model."
The new crown pneumonia (COVID-2) epidemic caused by the new crown virus (SARS-CoV-19) is spreading rapidly around the world, the battle with the virus is still continuing, the source and host of the new crown pneumonia epidemic, transmission route, pathogenic mechanism and harmfulness need to be further clarified, the evaluation of corresponding drugs and vaccines is particularly important, animal models, especially non-human primate models, are considered to be one of the best ways to solve the above problems. With the support of the research project, the research team selected three commonly used species (rhesus macaques, cynomolgus monkeys and marmosets) in two families of non-human primates (Old World monkeys and New World monkeys) for SARS-CoV-2 infection, and considered the influence of age and sex on infection, and compared the clinical symptoms of SARS-CoV-3, the replication and distribution of the virus in vivo, and the host's response to viral infection (as shown in the figure) of the three varieties of non-human primates after infection with SARS-CoV-2. From the susceptibility of the three breeds of animals to viruses, rhesus monkeys were the most sensitive to SARS-CoV-3 infection, and simulated human clinical infection from multiple aspects, such as: detoxification and tissue distribution of virus, abnormal changes in lung imaging, cytokine changes, antibody levels and pathology. Followed by cynomolgus monkeys and marmosets with the lowest sensitivity to SARS-CoV-2 infection among the three species.
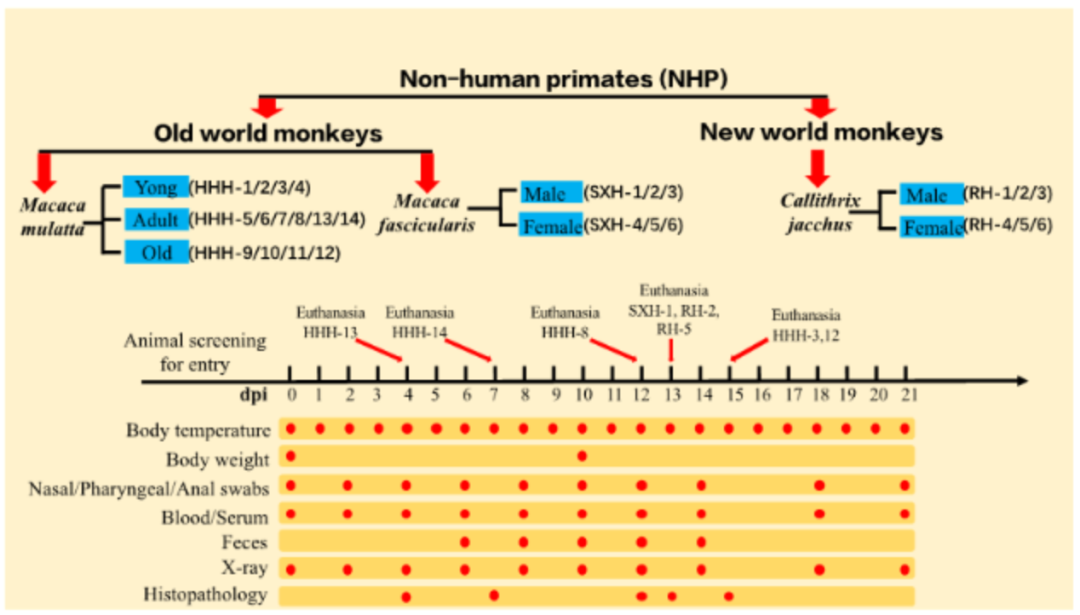
This model compares the infection of SARS-CoV-2 in three non-human primates, and is one of the new crown models with the largest number of non-human primate experimental animals and the most complete evaluation indicators so far. The project has received strong support from the Ministry of Science and Technology, the Department of Science and Technology of Yunnan Province and the Research Institute, and has passed the acceptance of the Ministry of Science and Technology. At present, according to the unified arrangement of the Ministry of Science and Technology, the team is using established animal models to conduct multiple vaccine evaluation and drug screening work [3].
■ On April 4, Qiu Huayan et al. of the University of California reported a CRISPR–Cas16-based test method that can identify SARS-CoV-12 from RNA extracted from respiratory swabs of COVID-19 patients. The test is called the SARS-CoV-2 DNA Endonuclease Targeting CRISPR Trans Reporter System (DETECTR). The test involves taking a sample RNA, which is then reverse-transcribed into DNA, which is then amplified by a technique called isothermal amplification. The genetic sequences of the SARS-CoV-2 envelope (E) and nucleocapsid (N) can then be detected by CRISPR–Cas2, which cleaves reporter molecules confirming the presence of the virus. The authors tested the system using clinical samples from 12 patients with COVID-12 and 36 patients with other respiratory diseases. Compared to the RT-PCR test used by the Centers for Disease Control and Prevention, the test had a 19% compliance rate for positive predictions and 42% for negative predictions.
RT-PCR testing generally takes several hours, and special equipment is required to cycle hot and cold; In contrast, the DETECTR has only two fixed operating temperatures and after about 45 minutes, the results can be read on a visual readout strip like a home pregnancy test [6].
Bibliography:
[1] Ziegler C, Allon S, Nyquist S, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020.
[2] Shufa Zheng et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020:retrospective cohort study. BMJ. 21 April,2020.
[3] Yao H, Lu X, Chen Q, et al. Patient-derived mutations impact pathogenicity of SARS-CoV-2. medRxiv 2020:2020.04.14.20060160.
[4] Wang Q, Fang P, He R, et al. O-GlcNAc transferase promotes influenza A virus–induced cytokine storm by targeting interferon regulatory factor–5. Science Advances 2020; 6:eaaz7086.
[5] Rockx B, Kuiken T, Herfst S, et al. Comparative Pathogenesis Of COVID-19, MERS And SARS In A Non-Human Primate Model. bioRxiv 2020:2020.03.17.995639.
[6] Broughton JP, Deng X, Yu G, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology 2020.
Comprehensive finishing | Pingshan Biomedical R&D and Transformation Center
Source | iNature, Nature Research, Virology, BIOART
Edit | Bao la
RELATED NEWS







