News attention
30
2020
-
12
Research Trends | Pingshan Biomedical R&D and Transformation Center has made new progress in the study of the activation mechanism of bacterial E3 enzymes
Guide
Recently, the Pingshan Biomedical R&D and Transformation Center of Shenzhen Bay Laboratory cooperated with the Shenzhen Graduate School of Peking University to publish a report entitled "Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial" in the sub-journal "Communications Biology" in "Nature" ubiquitin ligase IpaH9.8", introducing new advances in the study of the activation mechanism of bacterial E3 enzymes. In this study, the self-inhibitory mechanism of IpaH3.9 and the substrate-induced activation mechanism were elucidated by analyzing the crystal structure of the full-length protein of IpaH8.9 and its crystal structure with the substrate protein hGBP8 complex.
(Long press to identify the QR code below to read the paper)
Recently, the Pingshan Biomedical R&D and Transformation Center of Shenzhen Bay Laboratory cooperated with the Shenzhen Graduate School of Peking University to publish a report entitled "Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial" in the sub-journal "Communications Biology" in "Nature" ubiquitin ligase IpaH9.8", introducing new advances in the study of the activation mechanism of bacterial E3 enzymes. In this study, the self-inhibitory mechanism of IpaH3.9 and the substrate-induced activation mechanism were elucidated by analyzing the crystal structure of the full-length protein of IpaH8.9 and its crystal structure with the substrate protein hGBP8 complex.
The IpaH family protein is a class of E3 ubiquitin ligases derived from gram-negative bacteria, which consists of three parts: the N-terminal T3SS signal sequence, the LRR domain responsible for binding substrates, and the C-terminal conserved NEL domain with E3 enzyme function. Such E3 enzymes promote the infectious process by hijacking the host's ubiquitin-proteasome signaling pathway to inhibit host inflammation and endogenous immune responses. In host cells, IpaH proteins are usually self-inhibited in the absence of substrates to avoid self-ubiquitination and degradation by the host's ubiquitin system, or to avoid the creation of free Ub chains that activate the host immune response. IpaH9.8 is an E3 enzyme secreted in Shigella that targets ubiquitination and degradation of proteins such as NEMO (NF-κB regulator) and GBP1 (guanylate-binding protein 1) in host cells, thereby affecting the host's immune response. In this study, the authors found and demonstrated that the Hydrophobic Cluster at the C-terminus of the LRR domain plays an important role in stabilizing the self-inhibitory state of IpaH9.8 by resolving the crystal structure of IpaH9.8 and combining serial mutations and in vitro ubiquitination experiments (Figure 1).
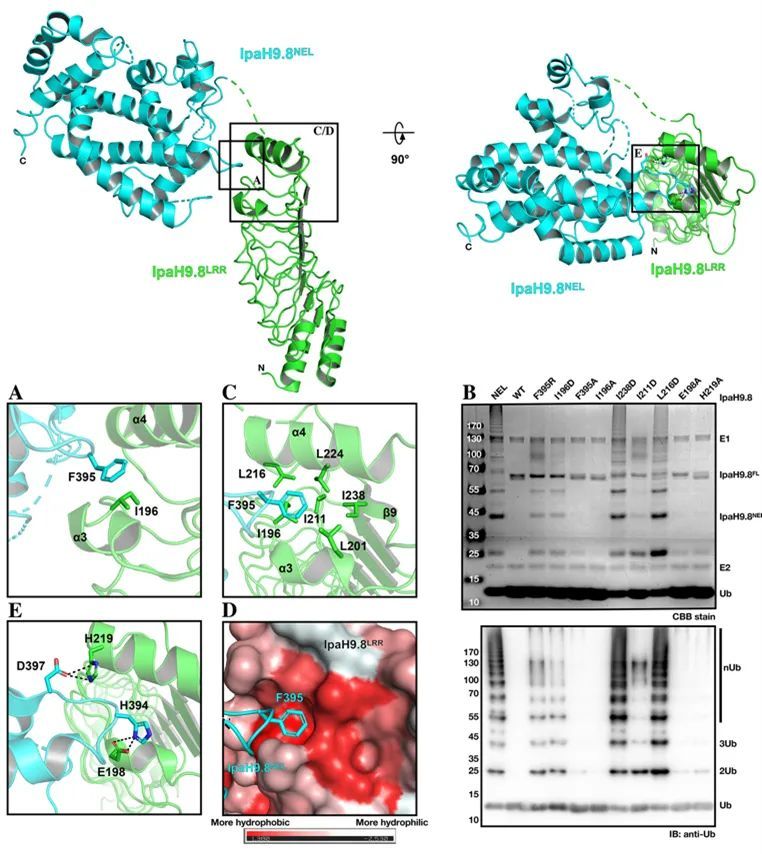
Figure 1 The Hydrophobic Cluster at the C-terminal of the IpaH9.8-LRR domain plays an important role in the self-inhibition of IpaH9.8
Subsequently, the authors resolved the LRR domain of IpaH9.8 and the complex crystal structure of the substrate protein hGBP1, and performed a comparative analysis with the structure of IpaH9.8 (Figure 2).
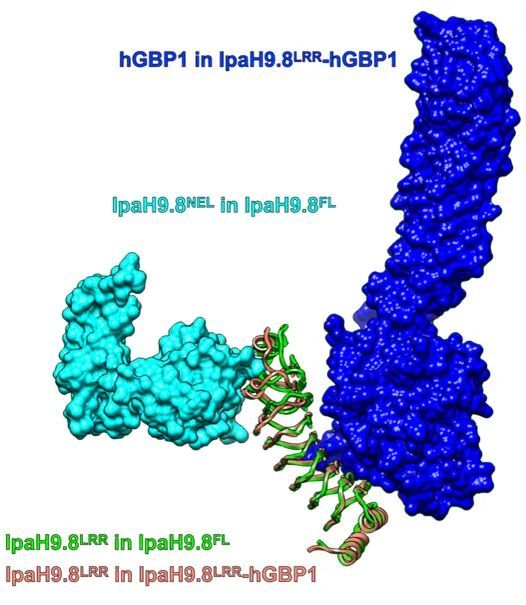
Fig. 2 Comparison of IpaH9.8LRR-hGBP1 complex structure with IpaH9.8 structure
By comparing the structure of the LRR domain of IpaH9.8 in different states, the authors found that the binding of substrates would cause significant conformational changes at the C-terminus of the LRR domain, which would destroy the stability of the Hydrophobic Cluster, thereby releasing the NEL domain and activating IpaH9.8. This view is confirmed by the normalized B-factor of the LRR domain in different states and the size of the hydrophobic cavity formed by the corresponding Hydrophobic Cluster (Figure 3).
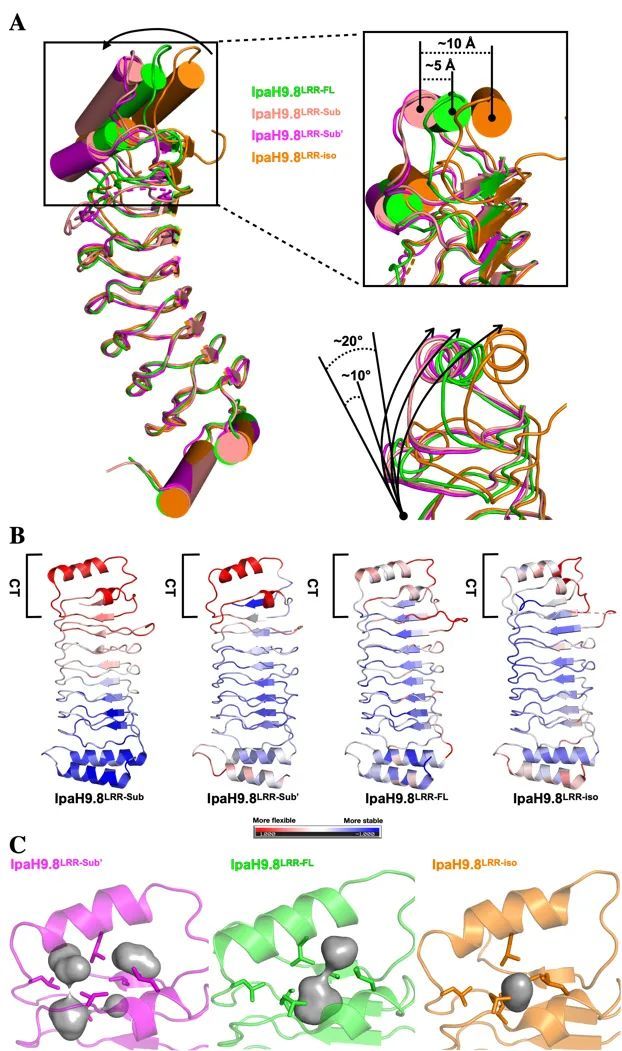
Figure 3 Substrate binding can unstabilize the Hydrophobic Cluster at the C-terminus of the IpaH9.8-LRR domain
In fact, the region of substrate binding on the LRR domain does not coincide with the Hydrophobic Cluster at the C-terminal (Figure 2), so why can substrate binding make the Hydrophobic Cluster at the C-terminal unstable? With this question in mind, the authors focused on the conformational changes of different regions at the C-terminus of the LRR domain after substrate binding, and found that the two amino acids, R166 and F187, also play the role of substrate sensors during substrate binding (Figure 4).
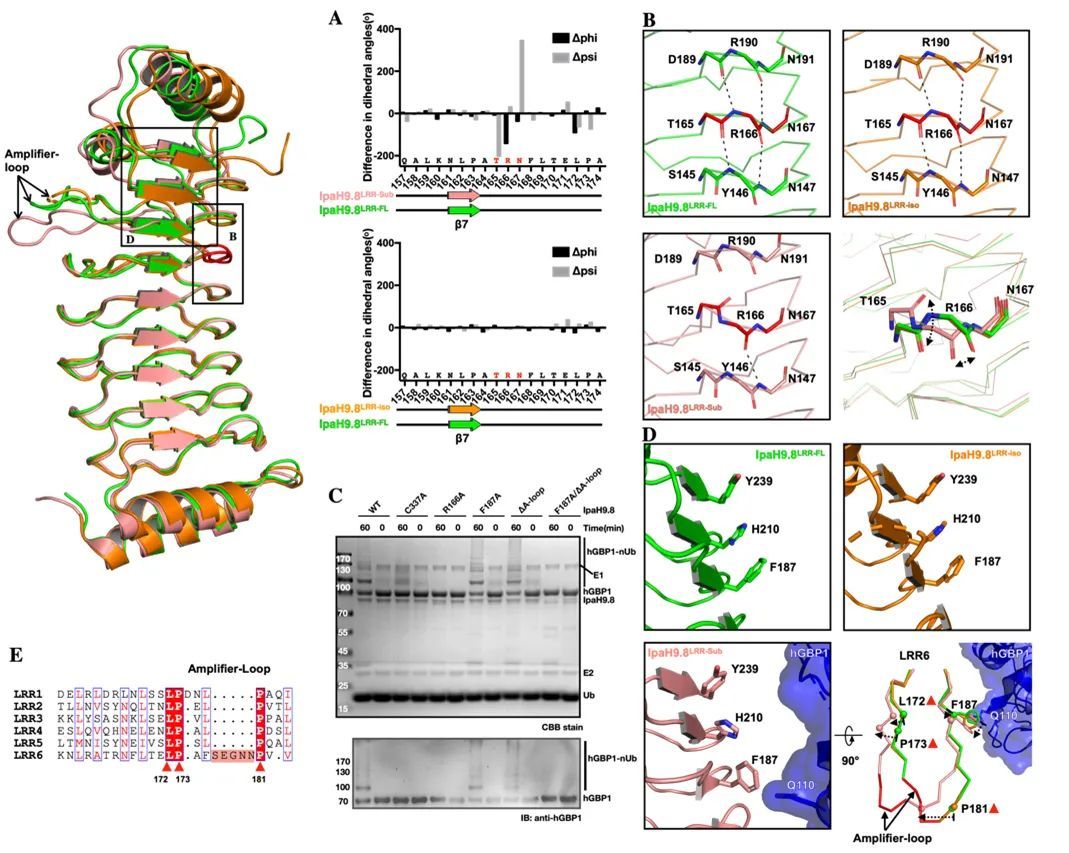
Figure 4 R9 and F8 located on the IpaH166.187-LRR domain can respond to the substrate arrival state of IpaH9.8 and act like a "molecular sensor"
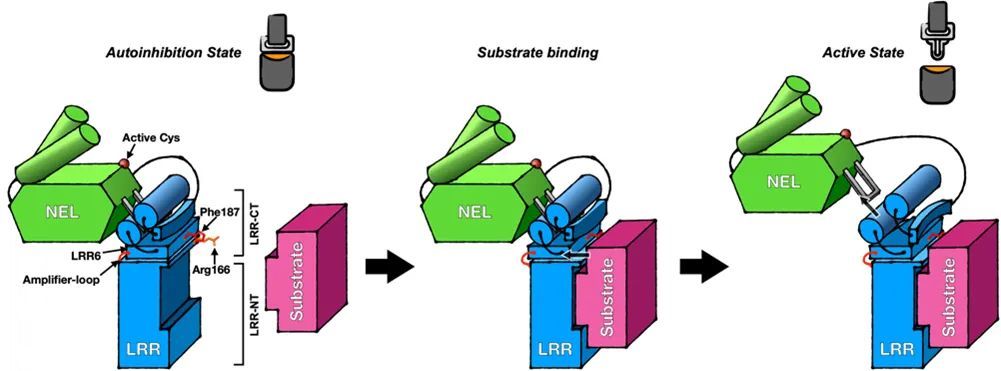
Figure 5 IpaH9.8 substrate binding-induced activation mechanism model
Dr. Yuxin Ye, Head of Protein Engineering Platform of the Department of Biology, Pingshan Biomedical R&D and Transformation Center, Shenzhen Bay Laboratory, and Hao Huang, Professor of the Laboratory of Structural Biology and Drug Development, Shenzhen Graduate School, Peking University, are co-corresponding authors of the paper. The research was supported by the National Natural Science Foundation of China, the Natural Science Foundation of Guangdong Province, and the Shenzhen Science and Technology Innovation Commission.
Edit | White
RELATED NEWS







